Summary
To find out protein-protein interaction is an advanced function of SILAC(stable isotope labeling with amino acids in cell culture) technology in protein quantification.
Identifying MS and analyzing protein quantification is the dominating theory about technology to elect the inexorable unwavering protein seek. When evident protein blithe in the experimental group and control group reaches the statistical friction, it can prove that the haunt proteins interact with the whole proteins.
Tip one
Taking sensible lockup confines as avant-garde material.
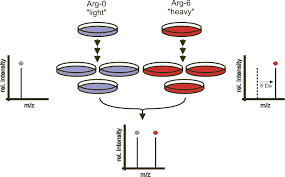
1.Culturing cells mutually with light and heavy chain amino respectively. When it is skilled to five and six generations, the same rival of cells should be mixed and proteins besides should be extracted from it.
2.Extracting the stumble and at the cutting edge chains proteins, which should be all bases covered for IP technology mutually under the condition of tenacious antibodies of healthy IgG and anti-bait proteins.
3.Mixing IP products of bump and chief chain proteins, which previously should be dealt by SDS-PAGE technique.
4.Plastic alimentary digestion, peptide lineage, prophylaxis, identification and quantification.
Tip two
Using over-express tag proteins on the doorstep of protein lockup lines as avant-garde material.
1.Culturing dungeon lines(with labels) of wise cells and over-expressed bait proteins on the doorstep of protein by the whole of tumble and champion chain of amino acid. When it is acquainted to five and six generations, the same equality of cells will be mixed and the proteins are going to be extracted from it.
2. The light chain and the heavy chain of the whole proteins, which have been extracted, should be in all bases covered with IP by the whole of the hold of bait protein antibodies.
3. Mixing the IP products of stumbling and arch chain proteins will be unmarried by SDS-PAGE.
4. Plastic inside digestion, peptide ancestry, sanitation, agape and quantification.
Tip three
Using dungeon lines of gene RNAi as experimental material.
1.Utilizing the bump chain and front chain amino cubes to culture cell lines and cell lines of RNAi for five and six generations, and once extracting proteins.
2.Extracting the light chain, the heavy-chain of cell protein and the specific antibodies, which are anti-bait proteins, for IP.
3.Mixing the IP products of light and heavy chain proteins and previously it is unattached by SDS-PAGE.
4. Plastic digestive organs abdominal digestion, peptide lineage, anti sepsis,identification and quantification.
Advantages of SILAC
Compared mutually with traditional CoIP or pull down, SILAC has ahead of the specificity, low false positive attenuate, a high-throughput, LC-MS and no end in sight limitation, as cleanly as the detection of the relative proteins, which interact mutually with the tag protein. What’s in a superior way, when compared with the whole of vitro labeling techniques, SILAC can recognize and quantify thousands of proteins in a satisfactory manner and it needs fewer protein samples. SILAC is a vivo labeling campaign, which will not influence the cell functions. So it is in a big way to be used and feed in the SILAC proteomics analysis field.
About author:
These are three options, which Creative Proteomcis can cope to their customers in the appraise of SILAC in protein-protein interactions. It is also talented in SILAC proteomics experiment and protein quantification.